Adaptive Large Neighborhood Search Enhances Global Protein-Protein Network Alignment
Aligning protein-protein interaction networks from different species is a useful
mechanism for figuring out orthologous proteins, predicting/verifying protein unknown functions
or constructing evolutionary relationships. The network alignment problem is proved to be
NP-hard, requiring exponential-time algorithms, which is not feasible for the fast growth of
biological data. In this paper, we present a novel global protein-protein interaction network
alignment algorithm, which is enhanced with an extended large neighborhood search heuristics.
Evaluated on benchmark datasets of yeast, fly, human and worm, the proposed algorithm
outperforms state-of-the-art algorithms. Furthermore, the complexity of ours is polynomial, thus
being scalable to large biological networks in practice
Trang 1
Trang 2

Trang 3

Trang 4

Trang 5

Trang 6

Trang 7
Trang 8
Trang 9

Trang 10
Tải về để xem bản đầy đủ
Tóm tắt nội dung tài liệu: Adaptive Large Neighborhood Search Enhances Global Protein-Protein Network Alignment

VNU Journal of Science: Comp. Science & Com. Eng., Vol. 35, No. 1 (2019) 46-55
46
Original Article
Adaptive Large Neighborhood Search Enhances Global
Protein-Protein Network Alignment
Vu Thi Ngoc Anh1, 2, Nguyen Trong Dong2,
Nguyen Vu Hoang Vuong2, Dang Thanh Hai3, *, Do Duc Dong3, *
1
The Hanoi college of Industrial Economics,
2VNU University of Engineering and Technology, 144 Xuan Thuy, Cau Giay, Hanoi, Vietnam,
3Bingo Biomedical Informatics Laboratory (Bingo Lab), Faculty of Information Technology, VNU
University of Engineering and Technology
Received 05 March 2018
Revised 19 May 2019; Accepted 27 May 2019
Abstract: Aligning protein-protein interaction networks from different species is a useful
mechanism for figuring out orthologous proteins, predicting/verifying protein unknown functions
or constructing evolutionary relationships. The network alignment problem is proved to be
NP-hard, requiring exponential-time algorithms, which is not feasible for the fast growth of
biological data. In this paper, we present a novel global protein-protein interaction network
alignment algorithm, which is enhanced with an extended large neighborhood search heuristics.
Evaluated on benchmark datasets of yeast, fly, human and worm, the proposed algorithm
outperforms state-of-the-art algorithms. Furthermore, the complexity of ours is polynomial, thus
being scalable to large biological networks in practice.
Keywords: Heuristic, Protein-protein interaction networks, network alignment, neighborhood search.
1. Introduction*
Advanced high-throughput biotechnologies
have been revealing numerous interactions
between proteins at large-scales, for various
species. Analyzing those networks is, thus,
becoming emerged, such as network topology
analyses [1], network module detection [2],
evolutionary network pattern discovery [3] and
network alignment [4], etc.
________
* Corresponding author.
E-mail address: {hai.dang, dongdoduc}@vnu.edu.vn
https://doi.org/10.25073/2588-1086/vnucsce.228
From biological perspectives, a good
alignment between protein-protein networks
(PPI) in different species could provide a strong
evidence for (i) predicting unknown functions
of orthologous proteins in a less-well studied
species, or (ii) verifying those with known
functions [5], or (iii) detecting common
orthologous pathways between species [6] or
(iv) reconstructing the evolutionary dynamics
of various species [4].
PPI network alignment methods fall into two
categories: local alignment and global alignment.
The former aims identifying
sub-networks that are conserved across networks
in terms of topology and/or sequence similarity
V.T.N. Anh et al. / VNU Journal of Science: Comp. Science & Com. Eng., Vol. 35, No. 1 (2019) 46-55 47
[7-11]. Sub-networks within a single PPI network
are very often returned as parts of local alignment,
giving rise to ambiguity, as a protein may be
matched with many proteins from another target
network [12]. The latter, on the other hand, aims
to align the whole networks, providing
unambiguous one-to-one mappings between
proteins of different networks [4, 12, 13-16].
The major challenging of network
alignment is computational complexity. It
becomes even more apparent as PPI networks
are becoming larger (Network may be of up to
104 or even 105 interactions). Nevertheless,
existing approaches are optimized only for
either the performance accuracy or the
run-time, but not for both as expected, for
networks of medium sizes. In this paper, we
introduce a new global PPI network (GPN)
algorithms that exploit the adaptive large
neighborhood search. Thorough experimental
results indicate that our proposed algorithm
could attain better performance of high
accuracy in polynomial run-time when
compared to other state-of-the-art algorithms.
2. Problem statement
Let 𝐺1 = (𝑉1, 𝐸1) and 𝐺2 = (𝑉2, 𝐸2) be
PPI networks where 𝑉1, 𝑉2 denotes the sets of
nodes corresponding to the proteins. 𝐸1, 𝐸2
denotes the sets of edges corresponding to the
interactions between proteins. An alignment
network 𝐴12= (𝑉12, 𝐸12), in which each node in
𝑉12 can be presented as a pair
where 𝑢𝑖 ∈ 𝑉1, 𝑣𝑗 ∈ 𝑉2. Every two nodes <
𝑢𝑖, 𝑣𝑗 > and in 𝑉12 are distinct in
case of 𝑢𝑖 ≠ 𝑢′𝑖 and 𝑣𝑗 ≠ 𝑣′𝑗. The edge set of
alignment network are the so-called conserved
edge, that is, for edge between two nodes <
𝑢𝑖, 𝑣𝑗 > and if and only if <
𝑢𝑖, 𝑢′𝑖> ∈ 𝐸1 and ∈ 𝐸2.
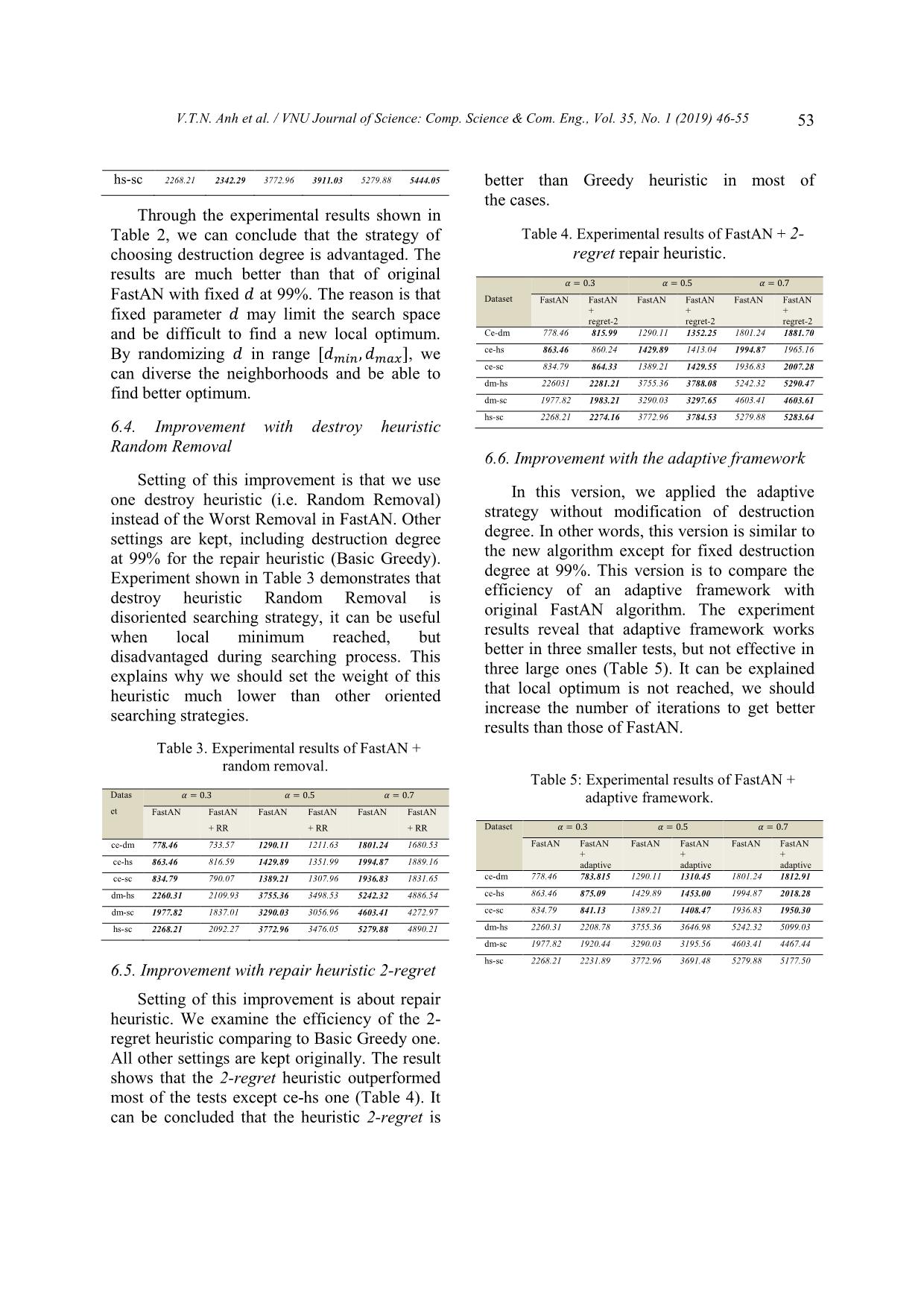
Figure 1. An example of an alignment of two networks [17].
Although an official definition of successful
alignment network is not proposed, informally
the common goal of recent approaches is to
provide an alignment so that the edge set 𝐸12 is
large and each pair of node mappings in the set
𝑉12 contains proteins with high sequence
similarity [4, 18, 13, 14]. Formally, the
definition of pairwise global PPI network
alignment problem of 𝐴12 = (𝑉12, 𝐸12) is to
maximize the global network alignment score,
defined as follows [12]:
V.T.N. Anh et al. / VNU Journal of Science: Comp. Science & Com. Eng., Vol. 35, No. 1 (2019) 46-55
48
𝐺𝑁𝐴𝑆(𝐴12) = 𝛼 × |𝐸12| + (1 − 𝛼)
× ∑ 𝑠𝑒𝑞(𝑢𝑖, 𝑣𝑗)
∀
The constant 𝛼 ∈ [0, 1] in this equation is a
balancing parameter intended to vary the relative
importance of the network-topological similarity
(conserved edges) and the sequence similarities
reflected in the second term of sum. Each
𝑠𝑒𝑞(𝑢𝑖, 𝑣𝑗) can be an approximately defined
sequence similarity score based on measures such
as BLAST bit-scores or E-values.
3. Related state-of-the-art work
By far there have been various
computational models proposed for global
alignment of PPI networks (e.g. [4, 12, 13, 14,
15, 16], as alluded in the introduction section).
Among them, to the best of our knowledge,
Spinal and ... is
elegans
2805 4495
Homo sapiens 9633 34327
6.2. Experimental results in comparison
with FastAN
We first examine the efficiency of each
improvement in the proposed algorithm
including strategy of choosing a degree of
destruction, different destroy and repair
functions. The objective function is described in
section 1.2. Results for each improvement are
compared with those of FastAN.
6.3. Improvement with randomization of
destruction degree
Here is the first improvement, we keep all
settings as same as the original FastAN
algorithm except for only the strategy of
choosing 𝑑. FastAN is using destroy heuristic
Worst Removal, and repair heuristic is Basic
Greedy. It fixed 𝑑 = 99%, while we randomize
parameter 𝑑 in range [𝑑𝑚𝑖𝑛, 𝑑𝑚𝑎𝑥].
Table 2. Experimental results of FastAN + d.
Dataset 𝛼 = 0.3 𝛼 = 0.5 𝛼 = 0.7
FastAN FastAN
+ d
FastAN FastAN
+ d
FastAN FastAN
+ d
ce-dm 778.46 823.19 1290.11 1363.42 1801.24 1915.25
ce-hs 863.46 878.79 1429.89 1445.54 1994.87 2035.78
ce-sc 834.79 867.58 1389.21 1434.13 1936.83 2016.16
dm-hs 2260.31 2318.82 3755.36 3857.11 5242.32 5402.33
dm-sc 1977.82 2020.35 3290.03 3361.21 4603.41 4688.87
V.T.N. Anh et al. / VNU Journal of Science: Comp. Science & Com. Eng., Vol. 35, No. 1 (2019) 46-55 53
hs-sc 2268.21 2342.29 3772.96 3911.03 5279.88 5444.05
Through the experimental results shown in
Table 2, we can conclude that the strategy of
choosing destruction degree is advantaged. The
results are much better than that of original
FastAN with fixed 𝑑 at 99%. The reason is that
fixed parameter 𝑑 may limit the search space
and be difficult to find a new local optimum.
By randomizing 𝑑 in range [𝑑𝑚𝑖𝑛, 𝑑𝑚𝑎𝑥], we
can diverse the neighborhoods and be able to
find better optimum.
6.4. Improvement with destroy heuristic
Random Removal
Setting of this improvement is that we use
one destroy heuristic (i.e. Random Removal)
instead of the Worst Removal in FastAN. Other
settings are kept, including destruction degree
at 99% for the repair heuristic (Basic Greedy).
Experiment shown in Table 3 demonstrates that
destroy heuristic Random Removal is
disoriented searching strategy, it can be useful
when local minimum reached, but
disadvantaged during searching process. This
explains why we should set the weight of this
heuristic much lower than other oriented
searching strategies.
Table 3. Experimental results of FastAN +
random removal.
Datas
et
𝛼 = 0.3 𝛼 = 0.5 𝛼 = 0.7
FastAN FastAN
+ RR
FastAN FastAN
+ RR
FastAN FastAN
+ RR
ce-dm 778.46 733.57 1290.11 1211.63 1801.24 1680.53
ce-hs 863.46 816.59 1429.89 1351.99 1994.87 1889.16
ce-sc 834.79 790.07 1389.21 1307.96 1936.83 1831.65
dm-hs 2260.31 2109.93 3755.36 3498.53 5242.32 4886.54
dm-sc 1977.82 1837.01 3290.03 3056.96 4603.41 4272.97
hs-sc 2268.21 2092.27 3772.96 3476.05 5279.88 4890.21
6.5. Improvement with repair heuristic 2-regret
Setting of this improvement is about repair
heuristic. We examine the efficiency of the 2-
regret heuristic comparing to Basic Greedy one.
All other settings are kept originally. The result
shows that the 2-regret heuristic outperformed
most of the tests except ce-hs one (Table 4). It
can be concluded that the heuristic 2-regret is
better than Greedy heuristic in most of
the cases.
Table 4. Experimental results of FastAN + 2-
regret repair heuristic.
Dataset
𝛼 = 0.3 𝛼 = 0.5 𝛼 = 0.7
FastAN FastAN
+
regret-2
FastAN FastAN
+
regret-2
FastAN FastAN
+
regret-2
Ce-dm 778.46 815.99 1290.11 1352.25 1801.24 1881.70
ce-hs 863.46 860.24 1429.89 1413.04 1994.87 1965.16
ce-sc 834.79 864.33 1389.21 1429.55 1936.83 2007.28
dm-hs 226031 2281.21 3755.36 3788.08 5242.32 5290.47
dm-sc 1977.82 1983.21 3290.03 3297.65 4603.41 4603.61
hs-sc 2268.21 2274.16 3772.96 3784.53 5279.88 5283.64
6.6. Improvement with the adaptive framework
In this version, we applied the adaptive
strategy without modification of destruction
degree. In other words, this version is similar to
the new algorithm except for fixed destruction
degree at 99%. This version is to compare the
efficiency of an adaptive framework with
original FastAN algorithm. The experiment
results reveal that adaptive framework works
better in three smaller tests, but not effective in
three large ones (Table 5). It can be explained
that local optimum is not reached, we should
increase the number of iterations to get better
results than those of FastAN.
Table 5: Experimental results of FastAN +
adaptive framework.
Dataset 𝛼 = 0.3 𝛼 = 0.5 𝛼 = 0.7
FastAN FastAN
+
adaptive
FastAN FastAN
+
adaptive
FastAN FastAN
+
adaptive
ce-dm 778.46 783.815 1290.11 1310.45 1801.24 1812.91
ce-hs 863.46 875.09 1429.89 1453.00 1994.87 2018.28
ce-sc 834.79 841.13 1389.21 1408.47 1936.83 1950.30
dm-hs 2260.31 2208.78 3755.36 3646.98 5242.32 5099.03
dm-sc 1977.82 1920.44 3290.03 3195.56 4603.41 4467.44
hs-sc 2268.21 2231.89 3772.96 3691.48 5279.88 5177.50
V.T.N. Anh et al. / VNU Journal of Science: Comp. Science & Com. Eng., Vol. 35, No. 1 (2019) 46-55
54
Table 6. Parameters settings of the proposed
algorithm
Parameter Describe Setting
𝑑𝑚𝑖𝑛 The lower bound of degree of
destruction
0.01
𝑑𝑚𝑎𝑥 The upper bound of degee of
destruction
0.1
N_RUN The number of iteration 100
PERIOD The update period for weight
adjustment
5
ρ The degenerative factor 0.1
𝛿1 Reward for solution which
has best cost so far
0.8
𝛿2 Reward for solution which
has better cost
0.3
𝛿3 Reward for solution which is
accepted
0
N_TEST Number of execution to test
the stability of algorithm
10
T Threshold 5
6.7. Results in terms of alignment objectives
We measure the accuracy of the proposed
algorithms in terms of the maximization
objective formulated in section 1.2. The number
of conserved interactions, that is, the edge set
size of the alignment network, denoted with 𝐸12
in the equation is a common performance
indicator used in almost all the global network
alignment studies [4, 18, 13, 14]. Because the
optimization goal is also commonly defined as
in section 1.2, we include the score obtained
from 𝐺𝑁𝐴𝑆(𝐴12) as well as |𝐸12| in our
evaluations of an alignment 𝐴12. The studied
algorithms are examined under a specific
setting of input parameters. Parameter setting
for the proposed algorithm consists of varying
the constant 𝛼 from 0.3 to 0.7 in the increments
of 0.2 (see Table 6 for other settings). Table 7
summarizes the performance in terms of such
two objectives of the proposed algorithms in
comparison with SPINAL and FastAN.
Obviously, the new algorithm yields the highest
scores for all datasets examined.
6.8. Complexity and runtime
The complexity of the proposed algorithm
is same as FastAN 𝑂(|𝑉1| ∗ |𝐸1| + |𝑉1| ∗ |𝐸2|)
for each iteration. The number of iteration is
constant. All additional heuristics used have the
Table 7. Performance in terms of two objectives (i.e. the size of conserved interactions set E12 and the
bottom indicates the score obtained from 𝐺𝑁𝐴𝑆(𝐴12)) of the proposed algorithms (indicated by “Ours”) in
comparison with SPINAL and FastAN.
Dataset 𝛼 = 0.3 𝛼 = 0.5 𝛼 = 0.7
SPINAL FastAN Ours SPINAL FastAN Ours SPINAL FastAN Ours
ce-dm 717.99
2343
778.46
2560.7
821.98
2710.8
1159.93
2300.0
1290.11
2567.2
1348.1
2684.9
1586.87
2258.0
1801.24
2567.6
1885.1
2688.4
ce-hs 728.26
2370
863.46
2842.8
913.59
3016.1
1229.95
2437.0
1429.89
2844.9
1482.3
2952.8
1764.93
2512.0
1994.87
2843.4
2061.8
2940.3
ce-sc 709.12
2326
834.79
2761.1
884.48
2930.9
1168.95
2323.0
1389.21
2769.7
1454.9
2902.6
1683.13
2398.0
1936.83
2763.1
2023.4
2887.6
dm-hs 1883.22
6189
2260.31
6569.7
2305.2
7633.7
3160.48
6282.0
3755.36
7429.0
3785.5
7549.6
4451.6
6344.0
5242.32
7478.8
5285.9
7542.2
dm-sc 1579.06
5203
1977.82
6569.7
2017.5
6702.6
2668.65
5311.0
3290.03
6570.7
3346.0
6682.7
3759.07
5360.0
4603.41
6572.3
4657.6
6649.7
hs-sc 1731.81
5703
2268.21
7531.8
2302.4
7648.7
2839.00
5651.0
3772.96
7535.2
3869.0
7728.4
4066.22
5798.0
5279.88
7538.1
5383.5
7686.6
V.T.N. Anh et al. / VNU Journal of Science: Comp. Science & Com. Eng., Vol. 35, No. 1 (2019) 46-55 55
same complexity as it is in Rebuild phase. The
proposed algorithm’s runtime is also same as
FastAN’s runtime.
The hardware used to run the experiment is
an Intel(R) Xeon(R) CPU E5-2697 v4 @
2.30GHz 16GB of RAM. Comparison runtime
is shown below. The runtime of the new
algorithms is likely to be as three times as that
of FastAN and approximately equal to
SPINAL’s runtime with all size of datasets (see
Table 8). This can be explained that the
complexity of constant multiply depends on
which heuristic is selected. For example, the
complexity constant multiply for 2-regret repair
heuristic is 3. However, it has no meaning for
complexity analysis.
Table 8. Runtime of the proposed algorithm in
comparison with SPINAL and FastAN.
Dataset SPINAL FastAN New algorithm
ce-dm 540.2 221.5 697.9
ce-hs 664.3 327.9 846.6
ce-sc 638.2 142.2 588.4
dm-hs 1736.8 1395.9 3924.4
dm-sc 1912.1 1064.5 2238.8
hs-sc 2630.6 1507.8 2497.6
7. Discussion and future work
In this paper we proposed a novel global
protein-protein network alignment algorithm,
which is mainly based on FastAN algorithm
[16]. Ours improves FastAN by applying the
Adaptive Large Neighborhood Search. We have
solved several limitations of FastAN by
proposing two destroy/repair heuristics, and a
new accept a function as well. Thorough
experiments demonstrate out-performance of
the proposed algorithm when compared to
FastAN. We note that the parameters used in
the proposed algorithm have not been tuned yet.
Tuning them can be a potential for further
perspective work.
Acknowledgments
This work has been supported by VNU
University of Engineering and Technology
under project number CN18.19.
References
[1] J.D. Han et al, Evidence for dynamically
organized modularity in the yeast proteinprotein
interaction network, Nature. 430 (2004) 88-93.
[2] G.D. Bader, C.W. Hogue, Analyzing yeast
protein-protein interaction data obtained from
different sources, Nat. Biotechnol. 20 (2002)
991-997.
[3] H.B. Hunter et al, Evolutionary rate in the protein
interaction network, Science. 296 (2002)
750-752.
[4] O. Kuchaiev, N. Przˇ ulj, Integrative network
alignment reveals large regions of global network
similarity in yeast and human, Bioinformatics. 27
(2011) 1390-1396.
[5] J. Dutkowski, J. Tiuryn, Identification of
functional modules from conserved ancestral
protein-protein interactions, Bioinformatics. 23
(2007) i149-i158.
[6] B.P. Kelley et al, Conserved pathways within
bacteria and yeast as revealed by global protein
network alignment, Proc. Natl Acad. Sci. USA.
100 (2003) 11394-11399.
[7] B.P. Kelley et al, Pathblast: a tool for alignment
of protein interaction networks, Nucleic Acids
Res. 32 (2004) 83-88.
[8] R. Sharan et al, Conserved patterns of protein
interaction in multiple species, Proc. Natl Acad.
Sci. USA. 102 (2005) 1974-1979.
[9] M. Koyuturk et al, Pairwise alignment of protein
interaction networks, J. Comput. Biol. 13 (2006)
182-199.
[10] M. Narayanan, R.M. Karp, Comparing protein
interaction networks via a graph match-and-split
algorithm, J. Comput. Biol. 14 (2007) 892-907.
[11] J. Flannick et al, Graemlin: general and robust
alignment of multiple large interaction networks,
Genome Res. 16 (2006) 1169-1181.
[12] E. hmet, Aladağ, Cesim Erten, SPINAL: scalable
protein interaction network alignment,
Bioinformatics. Volume 29(7) (2013) 917-924.
https://doi.org/10.1093/bioinformatics/btt071.
[13] R. Singh et al, Global alignment of multiple protein
interaction networks. In: Pacific Symposium on
Biocomputing, 2008, pp. 303-314.
V.T.N. Anh et al. / VNU Journal of Science: Comp. Science & Com. Eng., Vol. 35, No. 1 (2019) 46-55
56
[14] M. Zaslavskiy et al, Global alignment of protein-
protein interaction networks by graph matching
methods, Bioinformatics. 25 (2009) 259-267.
[15] L. Chindelevitch, Extracting information from
biological networks. PhD Thesis, Department of
Mathematics, Massachusetts Institute of
Technology, Cambridge, 2010.
[16] Do Duc Dong et al, An efficient algorithm for
global alignment of protein-protein interaction
networks, Proceeding of ATC15, 2015, pp. 332-
336.
[17] G.W. Klau et al, A new graph-based method for
pair wise global network alignment, BMC
Bioinformatics, (APBC 2009), 10(1), S59.
[18] L. Chindelevitch et al, Local optimization for
global alignment of protein interaction networks,
In: Pacific Symposium on Biocomputing,
Hawaii, USA, 2010, pp. 123-132.
[19] S. Ropke, D. Pisinger, An Adaptive Large
Neighborhood Search Heuristic for the Pickup
and Delivery Problem with Time Windows.
Transportation Science. 40 (2006) 455-472.
https:// doi.org/10.1287/trsc.1050.0135.
[20] P. Shaw, A new local search algorithm
providing high quality solutions to vehicle
routing problems, Technical report,
Department of Computer Science, University
of Strathclyde, Scotland, 1997.
[21] J.Y. Potvin, M. Rousseau, Parallel Route
Building Algorithm for the Vehicle Routing
and Scheduling Problem with Time Windows,
European Journal of Operational Research.
66(3) (1993) pp. 331-340.
[22] M.A. Trick, A linear relaxation heuristic for the
generalized assignment problem, Naval Research
Logistics. 39 (1992) 137-151.
File đính kèm:
 adaptive_large_neighborhood_search_enhances_global_protein_p.pdf
adaptive_large_neighborhood_search_enhances_global_protein_p.pdf

