Study on SARS-CoV-2 inhibition of some potential drugs using molecular docking simulation
Inhibitory capabilities of six old drugs selected from the DrugBank database including Losartan, Triazavirin, TMC-
310911, Verapamil, Clevudine and Elbasvir, which are promising for the treatment of an infectious disease caused by
SARS-CoV-2, were examined on the host receptor Angiotensin-converting enzyme 2 (ACE2) and the main protease
(PDB6LU7) of SARS-CoV-2 using molecular docking simulation. Results reveal that both proteins ACE2 and
PDB6LU7 are in strong inhibition by the drugs and the inhibitory effectiveness is in the order: Clevudine > Triazavirin
> TMC-310911 > Elbasvir > Losartan > Verapamil. In particular, the inhibitability highly correlates with the average
docking score energy of inhibitory complexes, and drug-protein active interactions. Regarding inhibitory ligands, their
polarizability, molecular size, and dispersion coefficient logP are also significant indicators for inhibition potential. The
drugs are suggested as valuable resources for selecting potential pharmaceuticals to prevent SARS-CoV-2 invasion into
human body given theoretical demonstration of molecular docking simulation.

Trang 1

Trang 2
Trang 3
Trang 4
Trang 5
Trang 6
Trang 7

Trang 8

Trang 9
Tóm tắt nội dung tài liệu: Study on SARS-CoV-2 inhibition of some potential drugs using molecular docking simulation

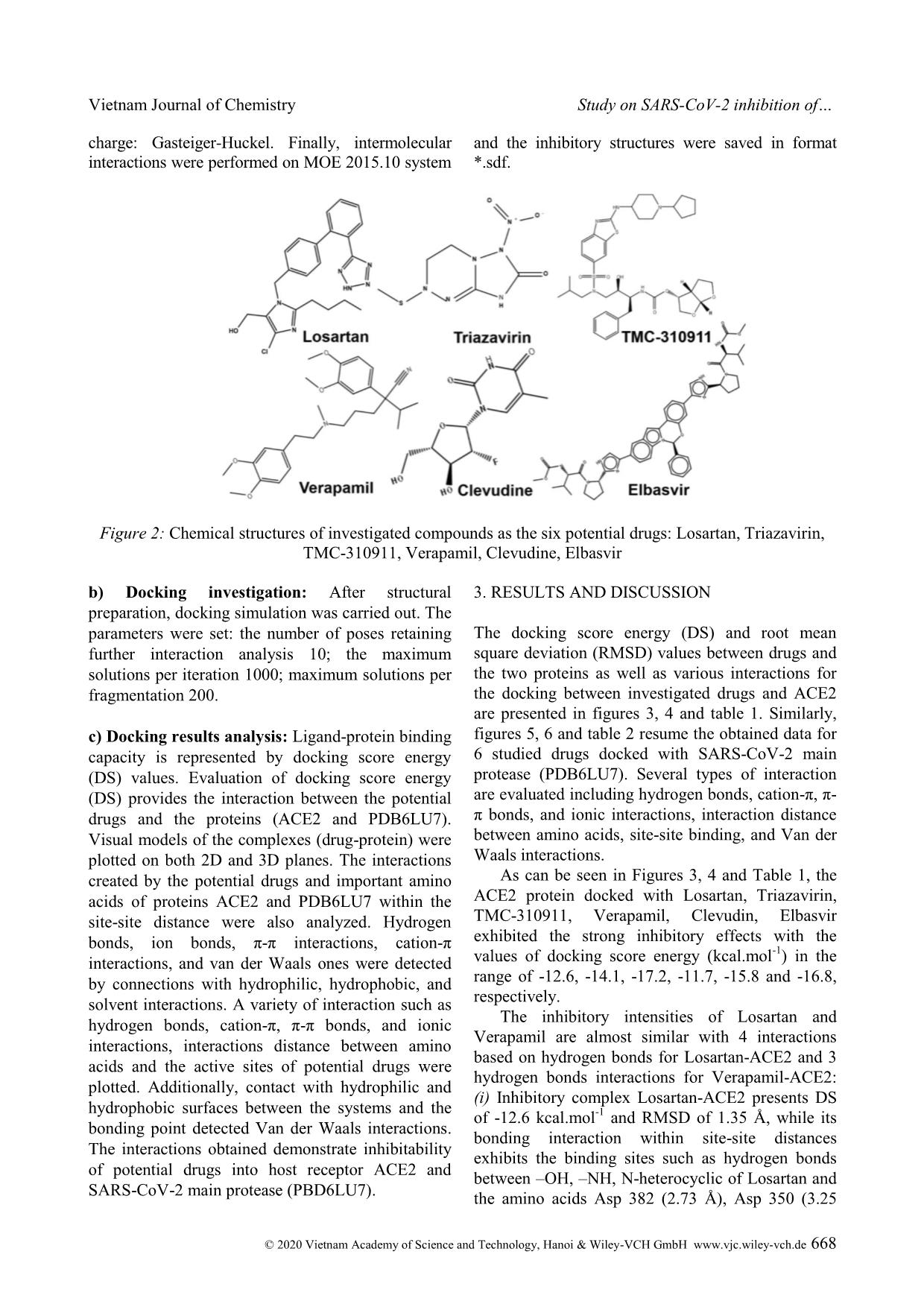
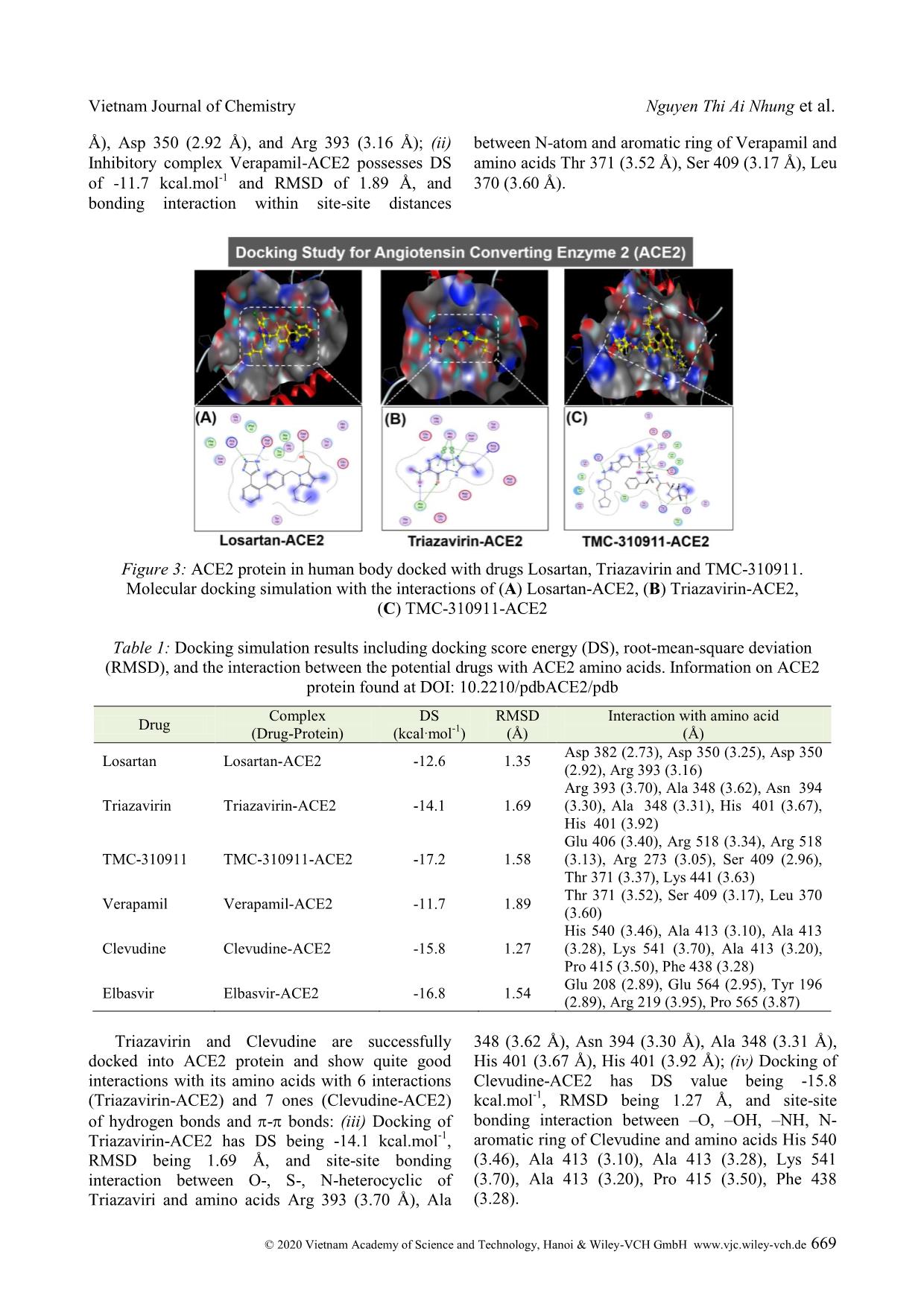
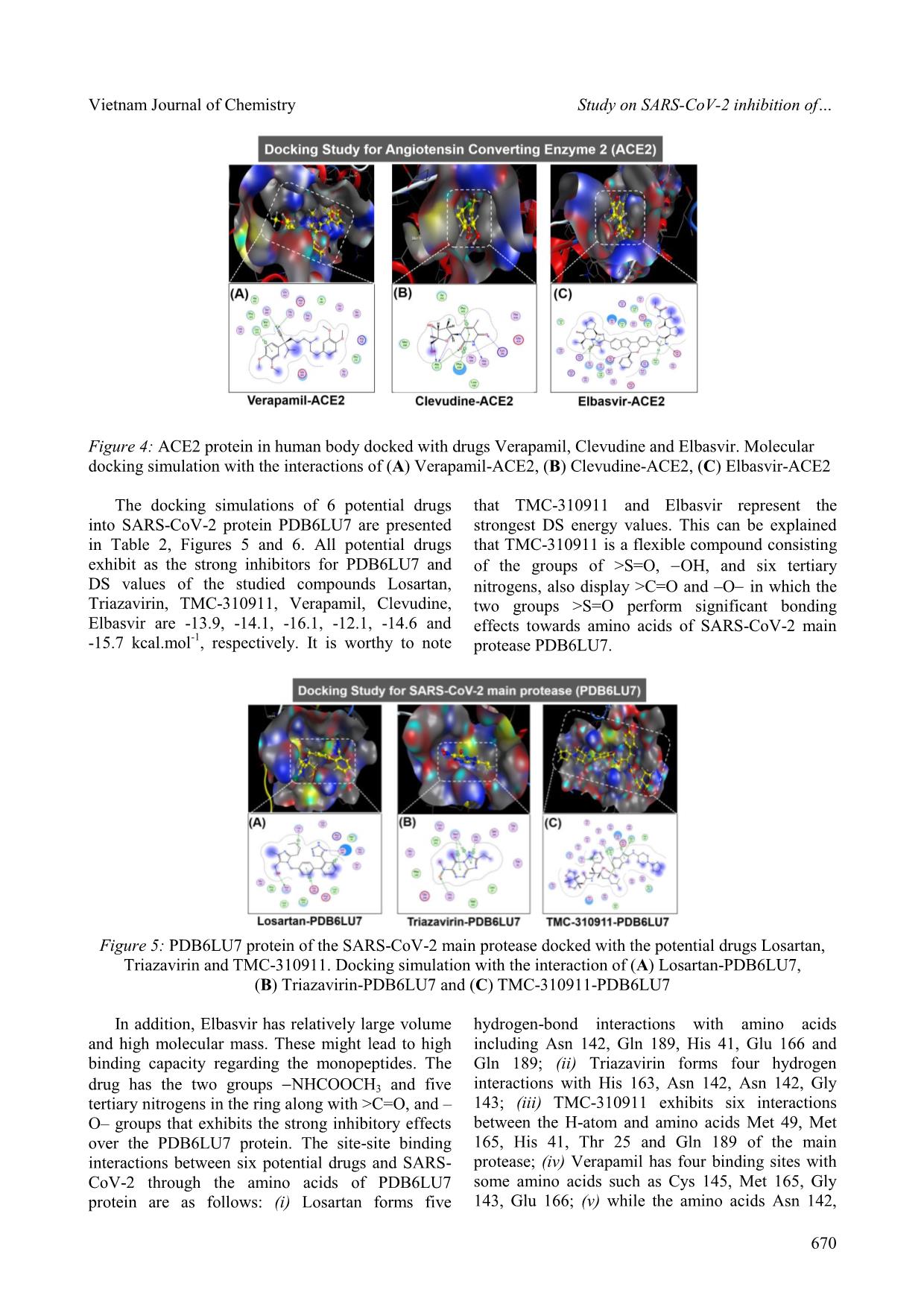
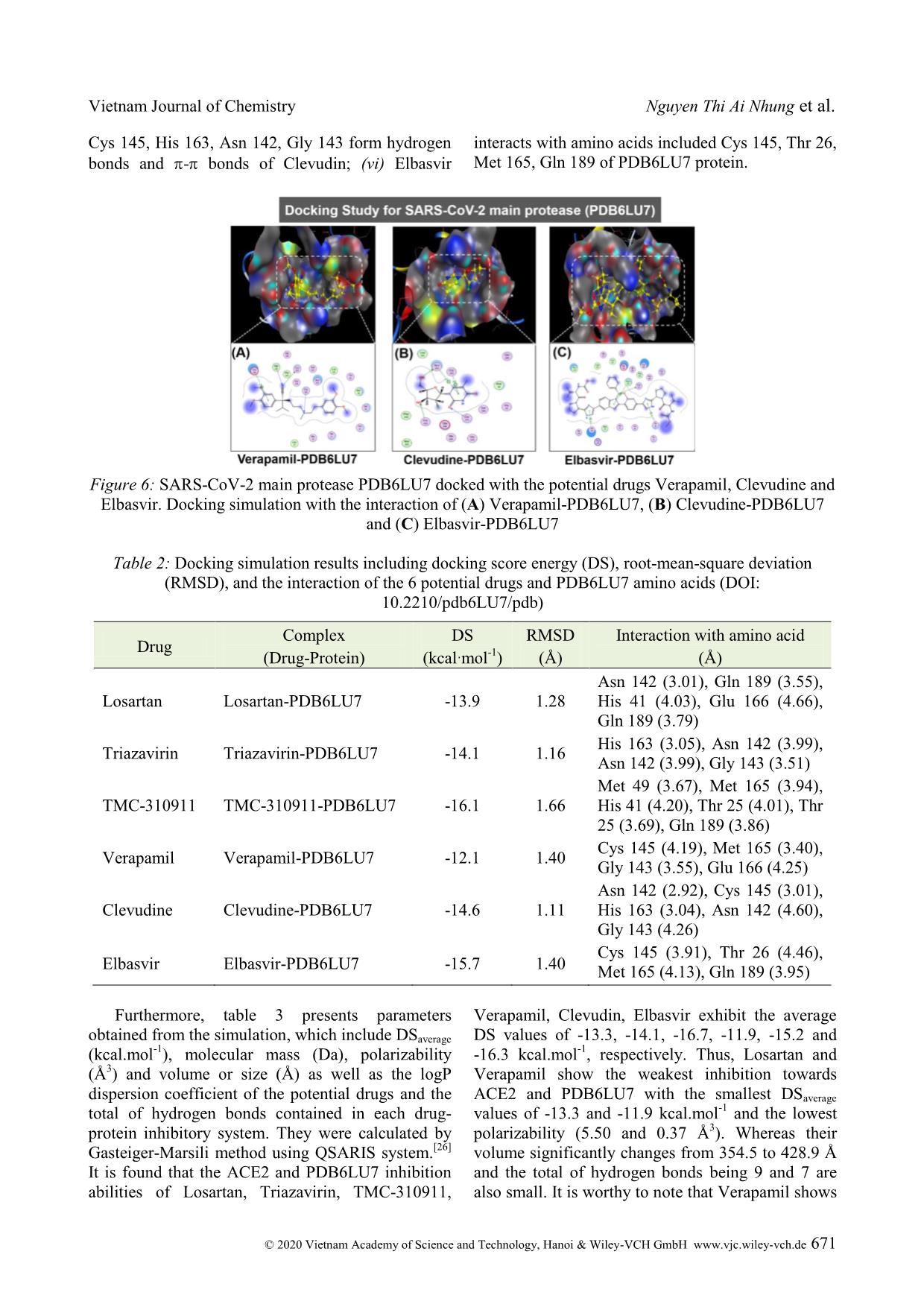
Cite this paper: Vietnam J. Chem., 2020, 58(5), 666-674 Article DOI: 10.1002/vjch.202000076 666 Wiley Online Library © 2020 Vietnam Academy of Science and Technology, Hanoi & Wiley-VCH GmbH Study on SARS-CoV-2 inhibition of some potential drugs using molecular docking simulation Tran Thi Ai My 1 , Le Trung Hieu 1 , Nguyen Thi Thanh Hai 1 , Huynh Thi Phuong Loan 1 , Thanh Q. Bui 1 , Bui Thi Phuong Thuy 2 , Phung Van Trung 3 , Bui Thi Bao Tram 4 , Nguyen Huyen Ngoc Tran 4 , Phan Tu Quy 5 , Duong Tuan Quang 6* , Doan Thi Yen Oanh 7 , Pham Van Tat 8 , Duy Quang Dao 9,10 , Nguyen Thi Ai Nhung 1* 1 Department of Chemistry, University of Sciences, Hue University, 77 Nguyen Hue, Hue, 53000, Viet Nam 2 Faculty of Fundamental Science, Van Lang University, 45 Nguyen Khac Nhu, district 1, Ho Chi Minh, 70000, Viet Nam 3 Center for Research and Technology Transfer, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet, Cau Giay district, Hanoi 10000, Viet Nam 4 Faculty of Medicine and Pharmacy, Van Lang University, Ho Chi Minh, 70000, Viet Nam 5 Department of Natural Sciences and Technology, Tay Nguyen University, 567 Le Duan Street, Buon Ma Thuot, 63000, Viet Nam 6 University of Education, Hue University, 34 Le Loi, Hue, 53000, Viet Nam 7 Publishing House for Science and Technology, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet, Cau Giay, Hanoi 10000, Viet Nam 8 Institute of Development and Applied Economics, Hoa Sen University, Ho Chi Minh, 70000, Viet Nam 9 Institute of Research and Development, Duy Tan University, 254 Nguyen Van Linh, Da Nang, 55000, Viet Nam 10 Faculty of Environmental and Chemical Engineering, Duy Tan University, 254 Nguyen Van Linh, Da Nang, 55000, Viet Nam Received May 26, 2020; Accepted June 10, 2020 Abstract Inhibitory capabilities of six old drugs selected from the DrugBank database including Losartan, Triazavirin, TMC- 310911, Verapamil, Clevudine and Elbasvir, which are promising for the treatment of an infectious disease caused by SARS-CoV-2, were examined on the host receptor Angiotensin-converting enzyme 2 (ACE2) and the main protease (PDB6LU7) of SARS-CoV-2 using molecular docking simulation. Results reveal that both proteins ACE2 and PDB6LU7 are in strong inhibition by the drugs and the inhibitory effectiveness is in the order: Clevudine > Triazavirin > TMC-310911 > Elbasvir > Losartan > Verapamil. In particular, the inhibitability highly correlates with the average docking score energy of inhibitory complexes, and drug-protein active interactions. Regarding inhibitory ligands, their polarizability, molecular size, and dispersion coefficient logP are also significant indicators for inhibition potential. The drugs are suggested as valuable resources for selecting potential pharmaceuticals to prevent SARS-CoV-2 invasion into human body given theoretical demonstration of molecular docking simulation. Keywords. SARS-CoV-2, potential drugs, ACE2, PDB6LU7, docking simulation. 1. INTRODUCTION Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2) is a new strain of coronaviruses family. It causes human diseases such as respiratory, gastrointestinal and neurological disorders, assembling the symptoms of the known diseases including Middle East Respiratory Syndrome (MERS-CoV) and Severe Acute Respiratory Syndrome (SARS). [1-3] It has only been about 4 months since the World Health Organisation (WHO) officially announced the first cases of infection with Vietnam Journal of Chemistry Nguyen Thi Ai Nhung et al. © 2020 Vietnam Academy of Science and Technology, Hanoi & Wiley-VCH GmbH www.vjc.wiley-vch.de 667 SARS-CoV-2. They were recorded in Wuhan, Hubei, China. Nevertheless, there have been more than 3 million people worldwide confirmed positive with SARS-CoV-2 so far. Therefore, early finding of an effective treatment, conducive to the race of vaccine proliferation is of the goal to scientists. There have been hundreds of drugs applied to tackle acute respiratory illnesses, or infectious diseases. For example, Losartan is an angiotensin II receptor blocker (ARB), which is suggested by The Food and Drug Administration (FDA) of The United States to treat hypertension. [4] Espescially, it is similarly used for Angiotensin-converting enzyme (ACE) inhibitors when a patient has a cough. [4,5] Similarly, Verapamil is used to treat high blood pressure. [6] Clevudine, also known under the trade names Levovir and Revovir, is an antiviral drug for hepatitis B treatment in South Korea and the Philippines. Triazavirin has been proved performing high efficacy towards influenza A and B, including the highly developed H5N1 strain [7-10] in Russia. Because of the structural similarities between SARS-CoV-2 and H5N1, health officials are conducting studies on Triazavirin as a potential drug candidate to combat SARS-CoV-2. [8] TMC-310911 (known as ASC-09) is being investigated for fighting against various strains of HIV-1. [11] Like other anti-HIV drugs, TMC-310911 is also being screened as a promising candidate to tackle SARS- CoV-2 infection. [12] Elbasvir is a direct-acting antiviral drug used to treat infectious liver disease resulted from hepatitis C virus infection. [13] According to Balasubramaniam and Reis (2020), elbasvir was theoretically calculated to create the best and most stable binding with the three important proteins essential for SARS-CoV-2 replication. [14] However, there has not been any further official announcement about potential drugs to directly treat SARS-CoV-2 or its vaccine. Therefore, a study on the interaction mechanism between cytological receptors and potential drugs by docking simulation may initially provide useful information guiding preclinical experiments with a time-effecti ... x potential drugs and SARS- CoV-2 through the amino acids of PDB6LU7 protein are as follows: (i) Losartan forms five hydrogen-bond interactions with amino acids including Asn 142, Gln 189, His 41, Glu 166 and Gln 189; (ii) Triazavirin forms four hydrogen interactions with His 163, Asn 142, Asn 142, Gly 143; (iii) TMC-310911 exhibits six interactions between the H-atom and amino acids Met 49, Met 165, His 41, Thr 25 and Gln 189 of the main protease; (iv) Verapamil has four binding sites with some amino acids such as Cys 145, Met 165, Gly 143, Glu 166; (v) while the amino acids Asn 142, Vietnam Journal of Chemistry Nguyen Thi Ai Nhung et al. © 2020 Vietnam Academy of Science and Technology, Hanoi & Wiley-VCH GmbH www.vjc.wiley-vch.de 671 Cys 145, His 163, Asn 142, Gly 143 form hydrogen bonds and - bonds of Clevudin; (vi) Elbasvir interacts with amino acids included Cys 145, Thr 26, Met 165, Gln 189 of PDB6LU7 protein. Figure 6: SARS-CoV-2 main protease PDB6LU7 docked with the potential drugs Verapamil, Clevudine and Elbasvir. Docking simulation with the interaction of (A) Verapamil-PDB6LU7, (B) Clevudine-PDB6LU7 and (C) Elbasvir-PDB6LU7 Table 2: Docking simulation results including docking score energy (DS), root-mean-square deviation (RMSD), and the interaction of the 6 potential drugs and PDB6LU7 amino acids (DOI: 10.2210/pdb6LU7/pdb) Drug Complex (Drug-Protein) DS (kcal·mol -1 ) RMSD (Å) Interaction with amino acid (Å) Losartan Losartan-PDB6LU7 -13.9 1.28 Asn 142 (3.01), Gln 189 (3.55), His 41 (4.03), Glu 166 (4.66), Gln 189 (3.79) Triazavirin Triazavirin-PDB6LU7 -14.1 1.16 His 163 (3.05), Asn 142 (3.99), Asn 142 (3.99), Gly 143 (3.51) TMC-310911 TMC-310911-PDB6LU7 -16.1 1.66 Met 49 (3.67), Met 165 (3.94), His 41 (4.20), Thr 25 (4.01), Thr 25 (3.69), Gln 189 (3.86) Verapamil Verapamil-PDB6LU7 -12.1 1.40 Cys 145 (4.19), Met 165 (3.40), Gly 143 (3.55), Glu 166 (4.25) Clevudine Clevudine-PDB6LU7 -14.6 1.11 Asn 142 (2.92), Cys 145 (3.01), His 163 (3.04), Asn 142 (4.60), Gly 143 (4.26) Elbasvir Elbasvir-PDB6LU7 -15.7 1.40 Cys 145 (3.91), Thr 26 (4.46), Met 165 (4.13), Gln 189 (3.95) Furthermore, table 3 presents parameters obtained from the simulation, which include DSaverage (kcal.mol -1 ), molecular mass (Da), polarizability (Å 3 ) and volume or size (Å) as well as the logP dispersion coefficient of the potential drugs and the total of hydrogen bonds contained in each drug- protein inhibitory system. They were calculated by Gasteiger-Marsili method using QSARIS system. [26] It is found that the ACE2 and PDB6LU7 inhibition abilities of Losartan, Triazavirin, TMC-310911, Verapamil, Clevudin, Elbasvir exhibit the average DS values of -13.3, -14.1, -16.7, -11.9, -15.2 and -16.3 kcal.mol -1 , respectively. Thus, Losartan and Verapamil show the weakest inhibition towards ACE2 and PDB6LU7 with the smallest DSaverage values of -13.3 and -11.9 kcal.mol -1 and the lowest polarizability (5.50 and 0.37 Å 3 ). Whereas their volume significantly changes from 354.5 to 428.9 Å and the total of hydrogen bonds being 9 and 7 are also small. It is worthy to note that Verapamil shows Vietnam Journal of Chemistry Study on SARS-CoV-2 inhibition of © 2020 Vietnam Academy of Science and Technology, Hanoi & Wiley-VCH GmbH www.vjc.wiley-vch.de 672 a LogP value of 5.7 being higher than 5, the value in which one observes weak inhibitory ability into the SARS-CoV-2. [26] Inversely, Clevudin, Triazavirin, TMC-310911 and Elbasvir display strong inhibition towards either ACE2 or PDB6LU7. The larger DSaverage values being -14.1 and -15.2 kcal.mol -1 exhibit for Clevudin and Triazavirin. Moreover, TMC-310911 and Elbasvir show the strongest inhibitory effects on two studied proteins with the largest DSaverage values of - 16.7 and -16.3 kcal.mol -1 by compared with the other drugs in the system. Additionally, the polarizability values of TMC-310911 and Elbasvir are significantly high (i.e. 8.80 and 9.97 Debye). Besides, their large molecular mass and volume might contribute to their high binding capacity with amino acids. However, it is noted that Clevudine and Triazavirin possess the smallest molecular mass and large polarizability values as well as strong site-site binding of interactions referred to hydrogen bonding. Based on these observations, it can be affirmed that Clevudine and Triazavirin bring their flexibilities and large number of interactions as well as impressive docking score energy values. They can be considered as the most potential candidates of six studied drugs. The results may satisfy for drugs screening requirements under the conditions of Lipinski's criteria: [27] (1) Molecular mass < 500 Da; (2) no more than 5 groups for hydrogen bonds; (3) no more than 10 groups receiving hydrogen bonds; (4) the value of logP is less than +5 (logP < 5). Based on the obtained results, we suggest the order of the potential drugs into the SARS-CoV-2 resistance as: Clevudine > Triazavirin > TMC- 310911 > Elbasvir > Losartan > Verapamil. Table 3: Molecular docking parameters including the average docking score energy values (DSaverage, kcal.mol -1 ), molecular mass (Da), polarizability (Å 3 ), volume or size (Å), logP and the total of hydrogen bonds of the 6 potential drugs docked with proteins ACE2 and PDB6LU7 Drug DSaverage Mass Polarizability Volume LogP Hydrogen bond Losartan -13.3 422.9 5.50 354.5 3.2 9 Triazavirin -14.1 232.2 7.88 165.8 1.1 11 TMC-310911 -16.7 756.0 8.80 542.0 3.7 13 Verapamil -11.9 454.6 0.37 428.9 5.7 7 Clevudine -15.2 260.2 9.39 211.7 1.2 12 Elbasvir -16.3 880.0 9.97 465.8 2.5 9 4. CONCLUSION Docking simulation results indicate that the selected drugs are potential inhibitors to host receptor ACE2 and SARS-CoV-2 main protease PDB6LU7. Clevudine and Triazavirin perform strongest inhibitability on both proteins. The respective inhibitability order of the active potential drugs towards both ACE2 and PDB6LU7 is summarized as: Clevudine > Triazavirin > TMC-310911 > Elbasvir > Losartan > Verapamil. Docking score energies of the drugs for inhibition of ACE2 vary from -17.2 to -11.7 kcal.mol -1 and the corresponding values for PDB6LU7 are from -12.1 to -16.1 kcal.mol -1 . The values calculated for root-mean-square deviation are lower than 2Å in all inhibitory systems. The potential drugs structurally are well-bound in their respective binding pose of the host receptor ACE2 and the targeted protease PDB6LU7. This is mainly based on hydrogen bond interactions between the studied compounds and in-pose amino acids. In particular, the inhibitability highly correlates with the average docking score energy of inhibitory complexes, and drug-protein active interactions. Regarding inhibitory ligands, their polarizability, molecular size, and dispersion coefficient logP are also significant indicators for inhibition potential. This study is highly conducive to further research in order to develop appropriate pharmaceuticals for clinical experiments on SARS-CoV-2. Acknowledgement. This research is supported by the Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 02/2020/ĐX. Conflict of interest. The authors declare no conflict of interest. REFERENCES 1. X. -W. Xu, X. -X. Wu, X. -G. Jiang, K. -J. Xu, L. -J. Ying, C. -L. Ma, S. -B. Li, H. -Y. Wang, S. Zhang, H.-N. Gao. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS- Cov-2) outside of Wuhan, China: retrospective case Vietnam Journal of Chemistry Nguyen Thi Ai Nhung et al. © 2020 Vietnam Academy of Science and Technology, Hanoi & Wiley-VCH GmbH www.vjc.wiley-vch.de 673 series, BMJ, 2020, 368. 2. L. Zou, F. Ruan, M. Huang, L. Liang, H. Huang, Z. Hong, J. Yu, M. Kang, Y. Song, J. Xia. SARS-CoV-2 viral load in upper respiratory specimens of infected patients, N. Engl. J. Med., 2020, 382, 1177-1179. 3. J. Parry. China coronavirus: cases surge as official admits human to human transmission, BMJ, 2020, m236. 4. M. McIntyre, S. Caffe, R. Michalak, J. Reid, Losartan. Losartan, an orally active angiotensin (AT1) receptor antagonist: a review of its efficacy and safety in essential hypertension, Pharmacol. Ther., 1997, 74, 181-194. 5. H. M. A. Abraham, C. M. White, W. B. White. The comparative efficacy and safety of the angiotensin receptor blockers in the management of hypertension and other cardiovascular diseases, Drug Saf., 2015, 38, 33-54. 6. G. Bakris, D. Sica, V. Ram, T. Fagan, P. T. Vaitkus, R. J. Anders. A comparative trial of controlled-onset, extended-release verapamil, enalapril, and losartan on blood pressure and heart rate changes, Am. J. Hypertens., 2002, 15, 53-57. 7. H. M. A. Abraham, C. M. White, W. B. White. Clevudine for the treatment of chronic hepatitis B virus infection, Expert Opin. Investig. Drugs, 2005, 14, 1277-1284. 8. M. J. Vincent, E. Bergeron, S. Benjannet, B. R. Erickson, P. E. Rollin, T. G. Ksiazek, N. G. Seidah, S. T. Nichol. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread, Virol. J., 2005, 2, 69. 9. A. V. Shvetsov, Y. A. Zabrodskaya, P. A. Nekrasov, V.V. Egorov. Triazavirine supramolecular complexes as modifiers of the peptide oligomeric structure, J. Biomol. Struct. Dyn., 2018, 36, 2694-2698. 10. R. M. Hoetelmans, I. Dierynck, I. Smyej, P. Meyvisch, B. Jacquemyn, K. Marien, K. Simmen, R. Verloes. Safety and pharmacokinetics of the HIV-1 protease inhibitor TMC310911 coadministered with ritonavir in healthy participants: results from 2 phase 1 studies, JAIDS J. Acquir. Immune Defic. Syndr., 2014, 65, 299-305. 11. H. -J. Stellbrink, K. Arastéh, D. Schürmann, C. Stephan, I. Dierynck, I. Smyej, R. M. Hoetelmans, C. Truyers, P. Meyvisch, B. Jacquemyn. Antiviral activity, pharmacokinetics, and safety of the HIV-1 protease inhibitor TMC310911, coadministered with Ritonavir, in treatment-naive HIV-1-infected patients, J. Acquir. Immune Defic. Syndr., 2014, 65, 283-289. 12. F. Mina, M. Rahman, S. Das, S. Karmakar, M. Billah, Potential drug candidates underway several registered clinical trials for battling COVID-19. 2020 13. S. Bagaglio, C. Uberti-Foppa, G. Morsica. Resistance mechanisms in hepatitis C virus: implications for direct-acting antiviral use, Drugs, 2017, 77, 1043- 1055. 14. M. Balasubramaniam, R. Shmookler Reis. Computational target-based drug repurposing of elbasvir, an antiviral drug predicted to bind multiple SARS-CoV-2 proteins, 2020. 15. S. R. Tipnis, N. M. Hooper, R. Hyde, E. Karran, G. Christie, A. J. Turner. A human homolog of angiotensin-converting enzyme cloning and functional expression as a captopril-insensitive carboxypeptidase, J. Bio. Chem., 2000, 275, 33238- 33243. 16. L. E. Gralinski, V. D. Menachery. Return of the Coronavirus: 2019-nCoV. Viruses, 2020, 12, 135. 17. D. Paraskevis, E. G. Kostaki, G. Magiorkinis, G. Panayiotakopoulos, G. Sourvinos, S. Tsiodras. Full- genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event, Infect. Genet. Evol., 2020, 79, 104212. 18. Homo Sapiens (Human). Https://www.Uniprot.Org/Uniprot/Q9BYF1 19. B. T. P. Thuy, T. T. A. My, N. T. T. Hai, L. T. Hieu, T. T. Hoa, H. Thi Phuong Loan, N. T. Triet, T. T. V. Anh, P. T. Quy, P. V. Tat. Investigation into SARS- CoV-2 resistance of compounds in garlic essential oil, ACS Omega, 2020. 20. The crytal structure of COVID-19 main protease in complex with an inhibitor N3. Https://www.wwpdb.org/pdb?id=pdb_00006lu7. Initial deposition on: 26 January 2020. 21. COVID information. https://www.drugbank.ca/covid- 19. 22. O. Tarasova, V. Poroikov, A. Veselovsky. Molecular docking studies of HIV-1 resistance to reverse transcriptase inhibitors: Mini-review, Molecules, 2018, 23, 1233. 23. T. M. C. Babu, S. S. Rajesh, B. V. Bhaskar, S. Devi, A. Rammohan, T. Sivaraman, W. Rajendra, Molecular docking, molecular dynamics simulation, biological evaluation and 2D QSAR analysis of flavonoids from Syzygium alternifolium as potent anti-Helicobacter pylori agents, RSC Adv., 2017, 7, 18277-18292. 24. T. -D. Ngo, T. -D. Tran, M. -T. Le, K. -M. Thai. Computational predictive models for P-glycoprotein inhibition of in-house chalcone derivatives and drug- bank compounds, Mol. Divers., 2016, 20, 945-961. 25. K. -M. Thai, D. -P. Le, T. -D. Tran, M. -T. Le. Computational assay of Zanamivir binding affinity with original and mutant influenza neuraminidase 9 Vietnam Journal of Chemistry Study on SARS-CoV-2 inhibition of © 2020 Vietnam Academy of Science and Technology, Hanoi & Wiley-VCH GmbH www.vjc.wiley-vch.de 674 using molecular docking, J. Theor. Biol., 2015, 385, 31-39. 26. J. Gasteiger, M. Marsili. Iterative partial equalization of orbital electronegativity-a rapid access to atomic charges, Tetrahedron, 1980, 36, 3219-3228. 27. C. A. Lipinski, F. Lombardo, B. W. Dominy, P. J. Feeney. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings, Adv. Drug Deliv. Rev., 1997, 23, 3-25. Corresponding authors: Duong Tuan Quang University of Education, Hue University 34, Le Loi street, Hue City 53000, Viet Nam E-mail: dtquang@hueuni.edu.vn orcid.org/0000-0002-4926-0271. Nguyen Thi Ai Nhung University of Sciences, Hue University 77, Nguyen Hue street, Hue City 53000, Viet Nam E-mail: ntanhung@hueuni.edu.vn orcid.org/0000-0002-5828-7898.
File đính kèm:
 study_on_sars_cov_2_inhibition_of_some_potential_drugs_using.pdf
study_on_sars_cov_2_inhibition_of_some_potential_drugs_using.pdf

