Secondary Metabolites from Marine Bacterium Nocardiopsis Sp. (G057)
Seven compounds were isolated and characterized from the culture broth of the marine bacteria
Nocardiopsis sp. (strain G057), which was isolated from sediment collecting at Cô Tô – Quảng Ninh. Their
structures were determined by spectroscopic analysis including MS and 2D NMR, as well as by comparison
with reported data in the literature. All compounds were evaluated for their antimicrobial activity against a
panel of clinically significant microorganisms. Compounds 1, 2 and 7 selectively inhibited Escherichia coli
with a MIC value of 32, 64, 8 μg/mL, respectively. Compound 3 exhibited antimicrobial activity against
several strains of both gram-positive and gram-negative bacteria

Trang 1

Trang 2
Trang 3
Trang 4

Trang 5
Bạn đang xem tài liệu "Secondary Metabolites from Marine Bacterium Nocardiopsis Sp. (G057)", để tải tài liệu gốc về máy hãy click vào nút Download ở trên
Tóm tắt nội dung tài liệu: Secondary Metabolites from Marine Bacterium Nocardiopsis Sp. (G057)

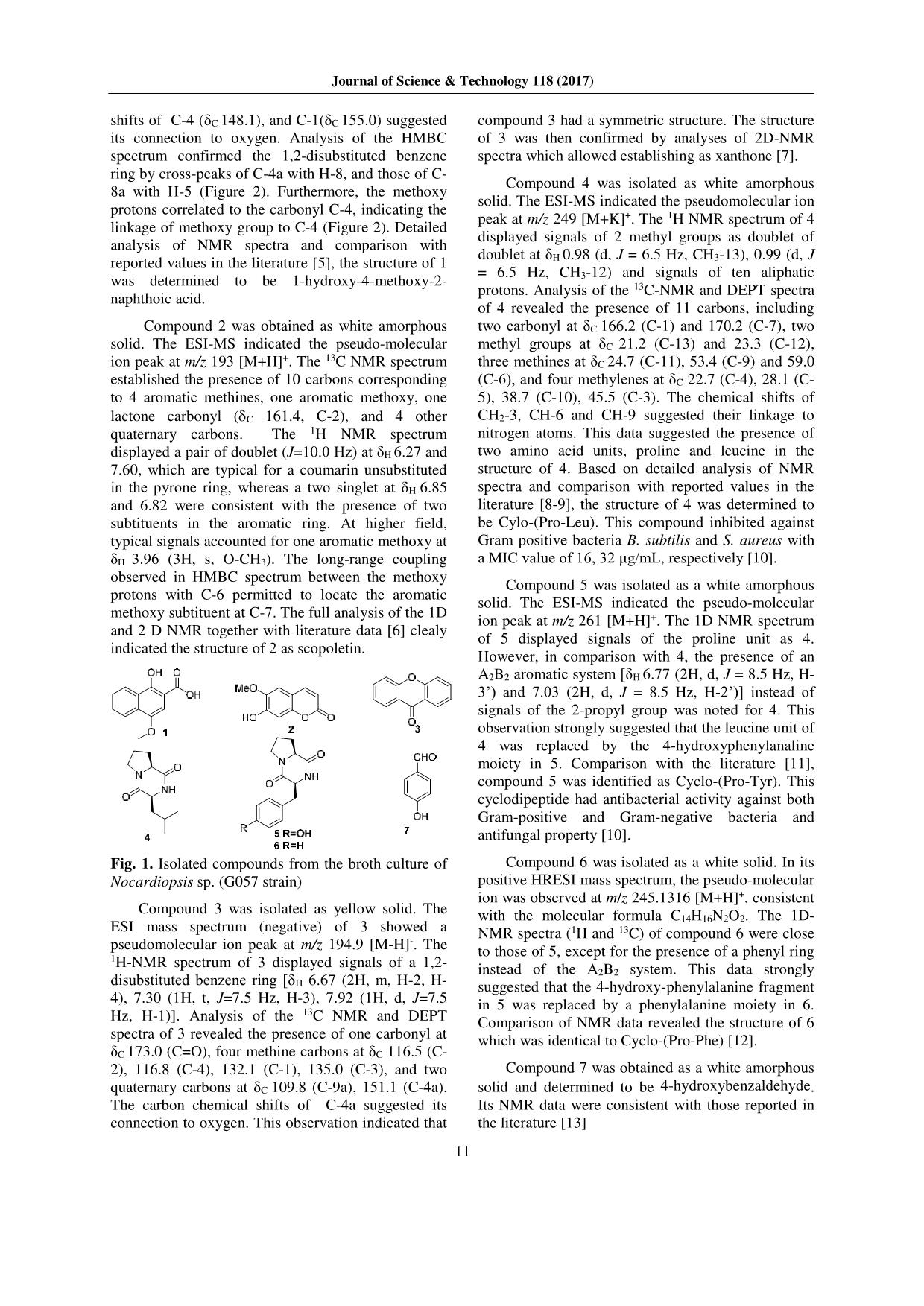
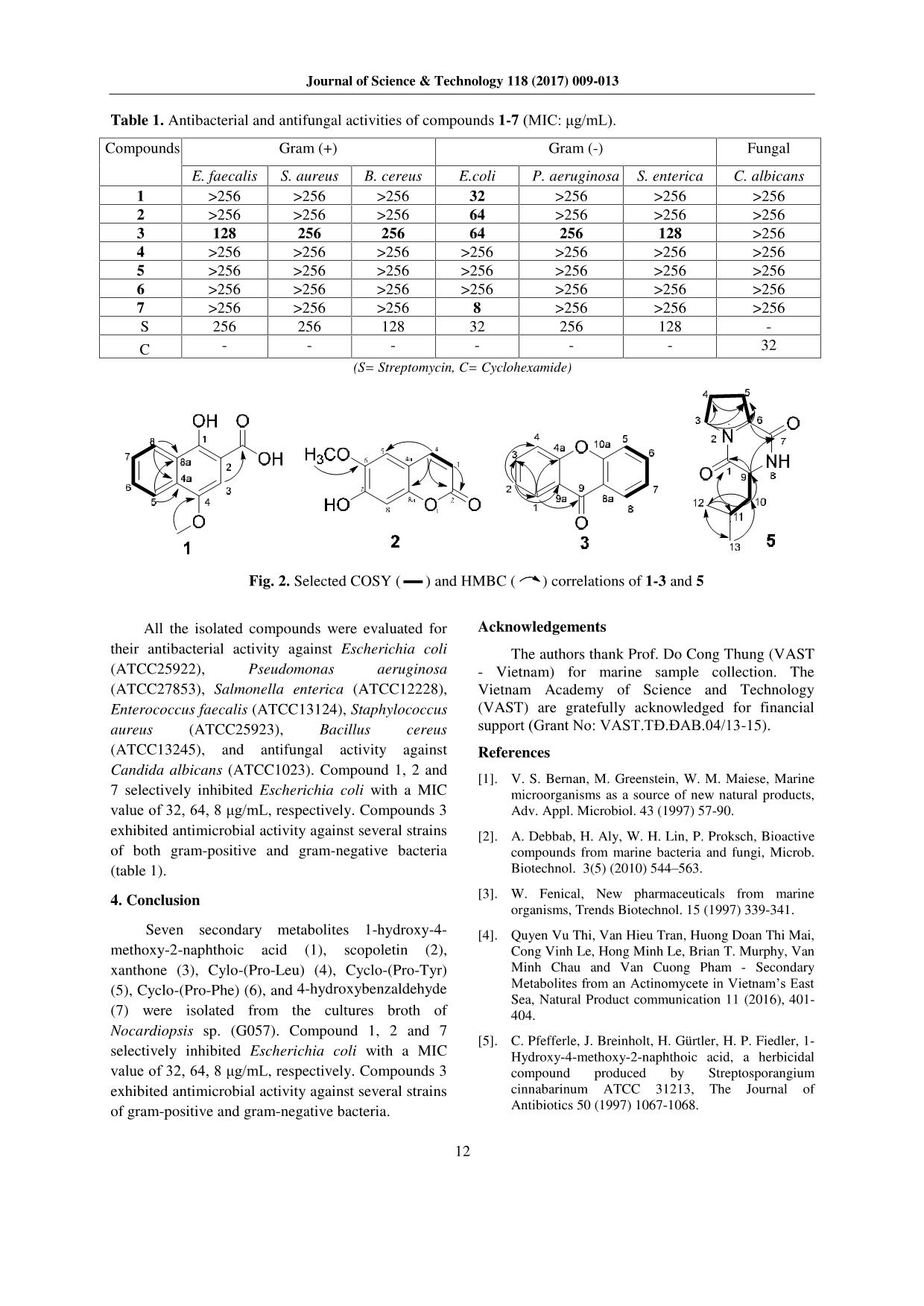
Journal of Science & Technology 118 (2017) 009-013 9 Secondary Metabolites from Marine Bacterium Nocardiopsis Sp. (G057) Tran Van Hieu1, Doan Thi Mai Huong1,*, Vu Thi Quyen1, Nguyen Quang Tung2, Chau Van Minh1, Pham Van Cuong1,* 1Institute of Marine Biochemistry-VAST, 18, Hoang Quoc Viet, Cau Giay, Hanoi, Viet Nam 2Hanoi University of Industry - Minh Khai Ward, Bac Tu Liem District, Hanoi, Vietnam Received: September 30, 2016; accepted: June 9, 2017 Abstract Seven compounds were isolated and characterized from the culture broth of the marine bacteria Nocardiopsis sp. (strain G057), which was isolated from sediment collecting at Cô Tô – Quảng Ninh. Their structures were determined by spectroscopic analysis including MS and 2D NMR, as well as by comparison with reported data in the literature. All compounds were evaluated for their antimicrobial activity against a panel of clinically significant microorganisms. Compounds 1, 2 and 7 selectively inhibited Escherichia coli with a MIC value of 32, 64, 8 μg/mL, respectively. Compound 3 exhibited antimicrobial activity against several strains of both gram-positive and gram-negative bacteria. Keywords: Nocardiopsis, marine microorganisms, antimicrobial activity, Cylo-(Leu-Pro), xanthone. 1. Introduction Marin* microorganisms have been the important study in recent years because of production of novel metabolites which represent various biological properties such as antiviral, antitumor or antimicrobial activities [1-2]. These secondary metabolites serve as model systems in discovery of new drugs [3]. In search of bioactive metabolites from marine bacteria, we examined the extract of the culture broth of the marine bacterium Nocardiopsis sp. (G057 strain). During our screening program, the EtOAc extract of this strain exhibited antimicrobial activity against both gram-positive (Enteroccocus faecalis - ATCC13124) and gram-negative (Escherichia coli - ATCC25922 and Salmonella enterica ATCC12228) bacteria strains, and the fungus strain (Candida albicans - ATCC1023). Herein, we described the isolation and structural determination of seven compounds (1 - 7) from the culture broth of Nocardiopsis sp. (G057) (Figure 1). Compound 1, 2 and 7 selectively inhibited Escherichia coli with a MIC value of 32, 64, 8 μg/mL, 3 exhibited antimicrobial activity against several strains of gram-positive and gram-negative bacteria. 2. Materials and Methods 2.1. General Experimental procedures ESI-MS were recorded on an Agilent 1100 LC- MSD Trap spectrometer. NMR spectra were recorded * Corresponding author: Tel: (+84) 4.37564995 Email:doanhuong7@yahoo.com on a Bruker 500.13 MHz spectrometer operating at 125.76 MHz for 13C NMR, and at 500.13 MHz for 1H NMR. 1H chemical shifts were referenced to CDCl3, DMSO-d6 and CD3OD at δ 7.27, 2.50 and 3.31 ppm, respectively, while the 13C chemical shifts were referenced to the central peak of at δ 77.1 (CDCl3), 39.5 (DMSO-d6), and 49.0 (CD3OD). For HMBC experiments the delay (1/2J) was 70 ms. TLC silica gel Merk 60 F254 was used as Thin-layer chromatography. Column chromatography (CC) was carried out using silica gel 40-63 µm or Sephadex LH-20. 2.2. Bacteria isolation and fermentation The marine sediment was collected from the coast of Cô Tô – Quảng Ninh in Vietnam in June of 2014. The sediment sample (1 g) was added to 10 mL of sterile sea water in a conical flask. The flask was agitated for about one hour. The marine sediment was filtered and the filtrate was serially diluted to obtain 10-1 to 10-7 dilutions using the sterilized sea water. An aliquot of 100 μL of each dilution was spread on the media. Different media like Starch Casein Agar (SCA), Glycerol Asparagine Agar (GA Agar), humic acid-B vitamin agar (HV Agar) and Glucose yeast malt extract agar (GYM) were used for isolation of actinomycetes. The media containing 50% of sterile sea water were supplemented with rifampicin (5 μg/mL) and nystatin (25 μg/mL) (Himedia Mumbai) to inhibit bacterial and fungal contamination, respectively. The petriplates were incubated for up to 3 weeks at 28 °C. The isolated discrete colonies were observed and used for identification. The fermentation was carried out in a 5 L flask using a modification of the published method [4]. Journal of Science & Technology 118 (2017) 10 2.3. Extraction and isolation The culture broth (50 L) of Nocardiopsis sp. (G057 strain) was filtered, and then extracted with ethyl acetate (30 L x 5 times). The extract was concentrated in vacuo to give 3.5 g of ethyl acetate extract. The ethyl acetate extract (3.5 g) was fractionated by column chromatography (CC) on silica gel, eluted with CH2Cl2/MeOH gradient to give five fractions. Fraction F1 was subjected to Sephadex LH-20 CC (MeOH) to afford four subfractions, subfraction F1.2 was subjected to CC on silica gel, eluted with mixtures of CH2Cl2/acetone (9/1) to afford 7 (6 mg). Fraction F2 (700 mg) was chromatographed by CC on Sephadex LH-20 (MeOH/CH2Cl2: 9/1), providing 3 subfractions. Subfraction F2.2 (200 mg) was subjected to CC on Sephadex LH-20 (MeOH/CH2Cl2: 9.5/0.5) affording 4 (10 mg). Subfraction F2.3 (170 mg) was chromatographed on Sephadex LH-20 (MeOH/CH2Cl2: 9/1), giving 6 (3.5 mg). Fraction F3 (1.3 g) was separated by CC on Sephadex LH-20 (MeOH/CH2Cl2: 9/1), leading to five subfractions. Subfraction 2 (200 mg) was purified by CC on Sephadex LH-20 (MeOH/CH2Cl2: 9/1) followed by preparative TLC (CH2Cl2/EtOAc: 7/1) to furnish compounds 5 (3.0 mg). Subfraction F3.5 (250 mg) was separated by Sephadex LH-20 CC (MeOH/CH2Cl2: 9/1) to provide 1 (4.5 mg). Fraction F4 (500 mg) was subjected to Sephadex LH-20 CC, eluting with a mixture of MeOH/CH2Cl2: 7/3, to yield 4 subfractions. Subfraction F4.2 (250 mg) was chromatographed on silica gel column, eluted with a solvent gradient of CH2Cl2/MeOH to afford 2 (4 mg) and 3 (15 mg). 1-hydroxy-4-methoxy-2-naphthoic acid (1): White amorphous solid, ESI-MS: m/z 241.04 [M+Na]+; 1H- NMR (500 MHz, MeOD): δH (ppm) 3.98 (3H, s, CH3O-4), 7.29 (1H, s, H-3), 7.51 (1H, dt, J=1.5, 8.0 Hz, H-7), 7.56 (1H, dt, J=1.5, 8.0 Hz, H-6), 8.15 (1H, br d, J= 8.0 Hz, H-5), 8.29 (1H, br d, J= 8.0 Hz, H-8). 13C- NMR (125 MHz, MeOD): δC (ppm) 56.1 (4- OCH3), 104.5 (C-3), 110.2 (C-2), 122.6 (C-5), 124.2 (C-8), 126,4 (C-7), 127.0 (C-8a), 128.3 (C-6), 130.1 (C-4a), 148.1 (C-4), 155.0 (C-1), 176.3 (C=O). Scopoletin (2): White amorphous solid, ESI-MS: m/z 193 [M+H]+; 1H- NMR (500 MHz, CDCl3): δH (ppm) 3,96 (3H, s, O-CH3), 6.27 (1H, d, J=9.5 Hz, H-3), 6.85 (1H, s, H-5), 6.92 (1H, s, H-8), 7.60 (1H, d, J=9.5 Hz, H-4). 13C- NMR (125 MHz, CDCl3): δC (ppm) 56.4 (O-CH3), 103.2 (C-8), 107.5 (C-5), 111.5 (C-4a), 113.5 (C-3), 143.2 (C-4), 144.0 (C-6), 149.7 (C-7), 150.3 (C-8a), 161.4 (C-2). Xanthone (3): Yellow solid, ESI-MS: m/z 194.9 [M- H]-; 1H- NMR (500 MHz, CDCl3): δH (ppm) 6.67 (2H, m, H-2, H-4); 7.30 (1H, t, J=7.5 Hz, H-3), 7.92 (1H, d, J=7.5 Hz, H-1). 13C- NMR (125 MHz, CDCl3): δC (ppm) 109.8 (C-8a), 116.5 (C-2), 116.8 (C-4), 132.1 (C-1), 135.0 (C-3), 151.1 (C-4a), 173.0 (C=O). Cylo-(Leu-Pro) (4): White amorphous solid, m.p 147- 148oC, ESI-MS: m/z 249 [M+K]+. 1H NMR (500 MHz, CD3OD): 0.98 (3H, d, J= 6.5 Hz, CH3-13), 0.99 (3H, d, J= 6.5 Hz, CH3-12), 1.54 (1H, m, Ha-10), 1.91 (1H, m, H-11), 1.93 (1H, m, Ha-4), 1.98 (1H, m, Hb- 4), 2.07 (1H, m, Hb-10), 2.08 (1H, m, Ha-5), 2.34 (1H, m, Hb-5), 3.55 (2H, m, CH2-3), 4.14 (1H, m, H- 9), 4.28 (1H, t, J= 7.5 Hz, H-6). 13C NMR (125 MHz, CD3OD): 21.2 (C-13), 22.7 (C-4), 23.3 (C-12), 24.7 (C-11), 28.1 (C-5), 38.7 (C-10), 45.5 (C-3), 53.4 (C- 9), 59.0 (C-6), 166.2 (C-1), 170.2 (C-7). Cyclo-(Pro-Tyr) (5): White amorphous solid, m.p 156-158oC, ESI-MS: m/z 261 [M+H]+. 1H NMR (500 MHz, CDCl3): 1.87 (2H, m, CH2-10), 1.99 (1H, m, H- 5b), 2.31 (1H, m, H-5a), 2.79 (1H, dd, J = 9.5, 14.5 Hz, Hb-10), 3.44 (1H, dd, J = 9.5, 14.5 Hz, Ha-10), 3.53 (1H, m, Hb-3), 3.64 (1H, m, Ha-3), 4.06 (1H, dd, J = 1.5, 7.5 Hz, H-6), 4.22 (1H, dd, J = 2.5, 9.5 Hz), 6.07 (1H, s, NH), 6.77 (2H, d, J = 8.5 Hz, H-3’), 7.03 (2H, d, J = 8.5 Hz, H-2’). Cyclo-(Pro-Phe) (6): White solid, HRESI-MS: m/z 245.1316 [M+H]+ (Calcd. 245.1290 for C14H17N2O2); 1H-NMR (400 MHz, CD3OD): δH (ppm) 1.60 (2H, m, Ha-4, Ha-5), 1.91-2.07 (3H, m, Hb-4, Hb- 5, Ha-10), 2.64 (1H, m, Hb-10), 3.02 (1H, dd, J = 4.8, 13.6 Hz, Ha-3), 3.22 (1H, dd, J = 4.4, 13.6 Hz, Hb-3), 3.59 (1H, m, H-6), 4.22 (1H, t, J = 4.8 Hz, H-9), 7.20-7.33 (5H, aromatic); 13C-NMR (100 MHz, CD3OD): δC (ppm) 21.1 (C-4), 28.4 (C-5), 39.6 (C-10), 44.7 (C-3), 57.7 (C-9), 58.4 (C-6), 127.1-129.9 (CH-aromatic), 135.3 (C-1’), 166.0 (C=O), 170.0 (C=O). 4-hydroxybenzaldehyde (7): Amorphous solid, 1H- NMR (500 MHz, CDCl3): δH (ppm) 6.95 (2H, d, J= 8.5 Hz, H-3, H-5), 7.80 (2H, d, J= 8.5 Hz, H-2, H-6), 9.87 (1H, s, -CHO). 3. Results and discussion Compound 1 was isolated as white amorphous solid. The 1H NMR spectrum of 1 showed signals of a 1,2-disubstituted benzene ring [δH 87.51 (1H, dt, J=1.5, 8.0 Hz, H-7), 7.56 (1H, dt, J=1.5, 8.0 Hz, H- 6), 8.15 (1H, br d, J= 8.0 Hz, H-5), 8.29 (1H, br d, J= 8.0 Hz, H-8)], and a singlet aromatic proton at δH 7.29 (1H, s, H-3). Signal of a singlet methoxy at δH 3.98 (3H, s, CH3O-4) was also noted. Analysis of the 13C NMR and DEPT spectra of 1 revealed the presence of 12 carbons, including one methoxy group at δC 56.1 (4-OCH3), ten aromatic carbons (five methines and five quaternary carbons), and one carbonyl at δC 176.3 (C=O). The carbon chemical Journal of Science & Technology 118 (2017) 11 shifts of C-4 (δC 148.1), and C-1(δC 155.0) suggested its connection to oxygen. Analysis of the HMBC spectrum confirmed the 1,2-disubstituted benzene ring by cross-peaks of C-4a with H-8, and those of C- 8a with H-5 (Figure 2). Furthermore, the methoxy protons correlated to the carbonyl C-4, indicating the linkage of methoxy group to C-4 (Figure 2). Detailed analysis of NMR spectra and comparison with reported values in the literature [5], the structure of 1 was determined to be 1-hydroxy-4-methoxy-2- naphthoic acid. Compound 2 was obtained as white amorphous solid. The ESI-MS indicated the pseudo-molecular ion peak at m/z 193 [M+H]+. The 13C NMR spectrum established the presence of 10 carbons corresponding to 4 aromatic methines, one aromatic methoxy, one lactone carbonyl (C 161.4, C-2), and 4 other quaternary carbons. The 1H NMR spectrum displayed a pair of doublet (J=10.0 Hz) at δH 6.27 and 7.60, which are typical for a coumarin unsubstituted in the pyrone ring, whereas a two singlet at δH 6.85 and 6.82 were consistent with the presence of two subtituents in the aromatic ring. At higher field, typical signals accounted for one aromatic methoxy at δH 3.96 (3H, s, O-CH3). The long-range coupling observed in HMBC spectrum between the methoxy protons with C-6 permitted to locate the aromatic methoxy subtituent at C-7. The full analysis of the 1D and 2 D NMR together with literature data [6] clealy indicated the structure of 2 as scopoletin. Fig. 1. Isolated compounds from the broth culture of Nocardiopsis sp. (G057 strain) Compound 3 was isolated as yellow solid. The ESI mass spectrum (negative) of 3 showed a pseudomolecular ion peak at m/z 194.9 [M-H]-. The 1H-NMR spectrum of 3 displayed signals of a 1,2- disubstituted benzene ring [δH 6.67 (2H, m, H-2, H- 4), 7.30 (1H, t, J=7.5 Hz, H-3), 7.92 (1H, d, J=7.5 Hz, H-1)]. Analysis of the 13C NMR and DEPT spectra of 3 revealed the presence of one carbonyl at δC 173.0 (C=O), four methine carbons at δC 116.5 (C- 2), 116.8 (C-4), 132.1 (C-1), 135.0 (C-3), and two quaternary carbons at δC 109.8 (C-9a), 151.1 (C-4a). The carbon chemical shifts of C-4a suggested its connection to oxygen. This observation indicated that compound 3 had a symmetric structure. The structure of 3 was then confirmed by analyses of 2D-NMR spectra which allowed establishing as xanthone [7]. Compound 4 was isolated as white amorphous solid. The ESI-MS indicated the pseudomolecular ion peak at m/z 249 [M+K]+. The 1H NMR spectrum of 4 displayed signals of 2 methyl groups as doublet of doublet at δH 0.98 (d, J = 6.5 Hz, CH3-13), 0.99 (d, J = 6.5 Hz, CH3-12) and signals of ten aliphatic protons. Analysis of the 13C-NMR and DEPT spectra of 4 revealed the presence of 11 carbons, including two carbonyl at δC 166.2 (C-1) and 170.2 (C-7), two methyl groups at δC 21.2 (C-13) and 23.3 (C-12), three methines at δC 24.7 (C-11), 53.4 (C-9) and 59.0 (C-6), and four methylenes at δC 22.7 (C-4), 28.1 (C- 5), 38.7 (C-10), 45.5 (C-3). The chemical shifts of CH2-3, CH-6 and CH-9 suggested their linkage to nitrogen atoms. This data suggested the presence of two amino acid units, proline and leucine in the structure of 4. Based on detailed analysis of NMR spectra and comparison with reported values in the literature [8-9], the structure of 4 was determined to be Cylo-(Pro-Leu). This compound inhibited against Gram positive bacteria B. subtilis and S. aureus with a MIC value of 16, 32 μg/mL, respectively [10]. Compound 5 was isolated as a white amorphous solid. The ESI-MS indicated the pseudo-molecular ion peak at m/z 261 [M+H]+. The 1D NMR spectrum of 5 displayed signals of the proline unit as 4. However, in comparison with 4, the presence of an A2B2 aromatic system [δH 6.77 (2H, d, J = 8.5 Hz, H- 3’) and 7.03 (2H, d, J = 8.5 Hz, H-2’)] instead of signals of the 2-propyl group was noted for 4. This observation strongly suggested that the leucine unit of 4 was replaced by the 4-hydroxyphenylanaline moiety in 5. Comparison with the literature [11], compound 5 was identified as Cyclo-(Pro-Tyr). This cyclodipeptide had antibacterial activity against both Gram-positive and Gram-negative bacteria and antifungal property [10]. Compound 6 was isolated as a white solid. In its positive HRESI mass spectrum, the pseudo-molecular ion was observed at m/z 245.1316 [M+H]+, consistent with the molecular formula C14H16N2O2. The 1D- NMR spectra (1H and 13C) of compound 6 were close to those of 5, except for the presence of a phenyl ring instead of the A2B2 system. This data strongly suggested that the 4-hydroxy-phenylalanine fragment in 5 was replaced by a phenylalanine moiety in 6. Comparison of NMR data revealed the structure of 6 which was identical to Cyclo-(Pro-Phe) [12]. Compound 7 was obtained as a white amorphous solid and determined to be 4-hydroxybenzaldehyde. Its NMR data were consistent with those reported in the literature [13] Journal of Science & Technology 118 (2017) 009-013 12 Table 1. Antibacterial and antifungal activities of compounds 1-7 (MIC: μg/mL). Compounds Gram (+) Gram (-) Fungal E. faecalis S. aureus B. cereus E.coli P. aeruginosa S. enterica C. albicans 1 >256 >256 >256 32 >256 >256 >256 2 >256 >256 >256 64 >256 >256 >256 3 128 256 256 64 256 128 >256 4 >256 >256 >256 >256 >256 >256 >256 5 >256 >256 >256 >256 >256 >256 >256 6 >256 >256 >256 >256 >256 >256 >256 7 >256 >256 >256 8 >256 >256 >256 S 256 256 128 32 256 128 - C - - - - - - 32 (S= Streptomycin, C= Cyclohexamide) Fig. 2. Selected COSY ( ) and HMBC ( ) correlations of 1-3 and 5 All the isolated compounds were evaluated for their antibacterial activity against Escherichia coli (ATCC25922), Pseudomonas aeruginosa (ATCC27853), Salmonella enterica (ATCC12228), Enterococcus faecalis (ATCC13124), Staphylococcus aureus (ATCC25923), Bacillus cereus (ATCC13245), and antifungal activity against Candida albicans (ATCC1023). Compound 1, 2 and 7 selectively inhibited Escherichia coli with a MIC value of 32, 64, 8 μg/mL, respectively. Compounds 3 exhibited antimicrobial activity against several strains of both gram-positive and gram-negative bacteria (table 1). 4. Conclusion Seven secondary metabolites 1-hydroxy-4- methoxy-2-naphthoic acid (1), scopoletin (2), xanthone (3), Cylo-(Pro-Leu) (4), Cyclo-(Pro-Tyr) (5), Cyclo-(Pro-Phe) (6), and 4-hydroxybenzaldehyde (7) were isolated from the cultures broth of Nocardiopsis sp. (G057). Compound 1, 2 and 7 selectively inhibited Escherichia coli with a MIC value of 32, 64, 8 μg/mL, respectively. Compounds 3 exhibited antimicrobial activity against several strains of gram-positive and gram-negative bacteria. Acknowledgements The authors thank Prof. Do Cong Thung (VAST - Vietnam) for marine sample collection. The Vietnam Academy of Science and Technology (VAST) are gratefully acknowledged for financial support (Grant No: VAST.TĐ.ĐAB.04/13-15). References [1]. V. S. Bernan, M. Greenstein, W. M. Maiese, Marine microorganisms as a source of new natural products, Adv. Appl. Microbiol. 43 (1997) 57-90. [2]. A. Debbab, H. Aly, W. H. Lin, P. Proksch, Bioactive compounds from marine bacteria and fungi, Microb. Biotechnol. 3(5) (2010) 544–563. [3]. W. Fenical, New pharmaceuticals from marine organisms, Trends Biotechnol. 15 (1997) 339-341. [4]. Quyen Vu Thi, Van Hieu Tran, Huong Doan Thi Mai, Cong Vinh Le, Hong Minh Le, Brian T. Murphy, Van Minh Chau and Van Cuong Pham - Secondary Metabolites from an Actinomycete in Vietnam’s East Sea, Natural Product communication 11 (2016), 401- 404. [5]. C. Pfefferle, J. Breinholt, H. Gürtler, H. P. Fiedler, 1- Hydroxy-4-methoxy-2-naphthoic acid, a herbicidal compound produced by Streptosporangium cinnabarinum ATCC 31213, The Journal of Antibiotics 50 (1997) 1067-1068. Journal of Science & Technology 118 (2017) 13 [6]. D. Akhmad, K. Soleh, B. S. K. Leonardus, and M. S. Yana, Scopoletin, a coumarin derivative compound isolated from Macaranga gigantifolia Merr, Journal of Applied Pharmaceutical Science 2 (2012) 175-177. [7]. A. A. Vitale, G. P. Romanelli, J. C. Autino, A. B. Pomilio, A novel route for the preparation of xanthones and chromanones, Journal of Chemical Research. Synopses 2 (1994) 82-83. [8]. F. Fdhila, V. Vazquez, J. L. Sanchez, R. Riguera, DD-Diketopiperazines. Antibiotics active against Vibrio anguillarum isolated from marine bacteria associated with cultures of Pecten maximus, J. Nat. Prod. 66 (2003) 1299-1301. [9]. C. Y. Wang, L. Han, K. Kang, C. L. Shao, Y. X. Wei, C. J. Zheng and H. S. Guan, Secondary metabolites from green algae Ulva pertusa, Chem. Nat. Compd., 68 (2010) 828-830. [10]. K. Nishanth, C. Mohandas, B. Nambisan, D. R. Soban Kumar, R. S. Lankalapalli, Isolation of proline-based cyclic dipeptides from Bacillus sp. N strain associated with rhabitid entomopathogenic nematode and its antimicrobial properties, World J Microbiol Biotechnol 29 (2013) 355–364. [11]. M. Mitova, G. Tommonaro, U. Hentschel, W. E. G. Muller, S. De Rosa, Exocellular cyclic dipeptides from a Ruegeria strain associated with cell cultures of Suberites domuncula, Mar. Biotechnol. 6 (2004) 95- 103. [12]. G. Wang, S. Dai, M. Chen, H. Wu, L. Xie, X. Luo, X. Li, Two diketopiperazine cyclo(Pro-Phe) isomers from marine bacterium Bacillus subtilis sp. 13-2, Chem. Nat. Compd. 46 (2010) 583-585. [13]. G. Venkateswara, C. Sharlene, and T. Mukhopadhyay, Secondary metabolites from the flowers of Mimusops elengi Linn., Der Pharmacia Lettre 4 (2012) 1817-1820.
File đính kèm:
 secondary_metabolites_from_marine_bacterium_nocardiopsis_sp.pdf
secondary_metabolites_from_marine_bacterium_nocardiopsis_sp.pdf

